cyto-chromosome-vis v1.3.3
cyto-chromosome-vis
Interactive visual representation/ web visualization tool for chromosome ideograms
This is a web component built with D3.js to render chromosome representations in SVG. Each chromosome has interactive features such as clicking a band, picking a specific cyto-location (base pair), or removing a specific selection. A convenient API is included to integrate user actions on the chromosomes with other js components.
This can be used in plain JavaScript (as a global namespace object), in Angular.js (as a directive or a service), or in CommonJS environments such as Node.js.
Demo
A demo is available here: http://linjoey.github.io/cyto-chromosome-vis/
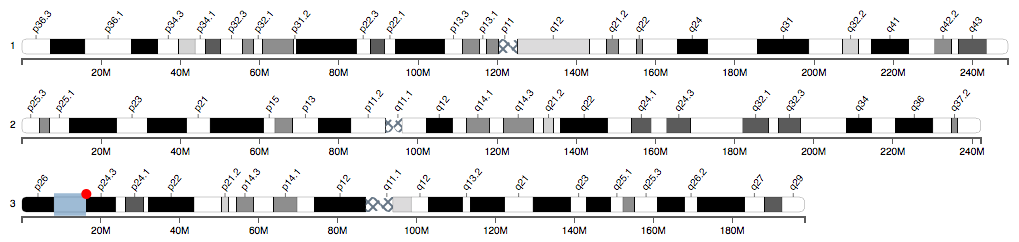
Usage
Install with npm: npm install --save cyto-chromosome-vis
Install with bower: bower install --save cyto-chromosome-vis
Use in plain JavaScript, Angular, or Node.js
Plain JavaScript
Include D3 and the project source:
cyto-chromosome.jsorcyto-chromosome.min.jsfrom the project root.Create a target div to host the chromosome
<div id="chrY"></div>- Instantiate a chromosome object.
var chromosomeFactory = cyto_chr.chromosome;
var x = chromosomeFactory()
.segment("X")
.target('#chrX')
.render();CommonJS
<div id="test"></div>
<script src="bundle.js"></script>/*src.js*/
var chromosomeFactory = require('../cyto-chromosome-vis').chromosome;
chromosomeFactory()
.target('#test')
.render();Here bundle.js is compiled with Browserify, e.g. browserify src.js > bundle.js
Angular.js
- Include the module
cyto-chromosome-visas a project dependency
angular.module('demoApp',['cyto-chromosome-vis'])- Simply use the directive
cytochromosome
<cytochromosome segment="X" width="1000" show-axis="true" use-relative="true" resolution="400"></cytochromosome>- If you require interacting with the chromosome instance from a controller, inject the service
cytochromosomeand instantiate manually:
<div id="chr3"></div>var app = angular.module('demoApp',['cyto-chromosome-vis'])
.config(['cytochromosomeProvider', function(cytochr) {
cytochr.setDataDir('data/');
}])
.controller('main', function($scope, cytochromosome) {
var c = cytochromosome.build()
.target("#chr3")
.segment(3)
.showAxis(true)
.render();
})If the data directory is moved from the default node_modules/cyto-chromosome-vis/data/, inject cytochromosomeProvider and configure the data path relative to the html page.
Once a chromosome is drawn, click a band to add a selector, alt/option click to add multiple selectors. Drag the edges of the selector to change the selection; click the red button on a selector to delete it.
Configurations API
Configure chromosomes with chromosome.config(type, value) or chromosome.type(value). chromosome.type() (no arguments) will return the current configuration. The default values for each configuration is shown below.
var s = cyto_chr.chromosome()
.config('segment', "2")
.config('target', '#chrX')
.config('resolution', "850") //400, 550, or 850
.config('width', 1200)
.config('useRelative', false)
.config('showAxis', false)
.render();
s.resolution(); //850
s.width(); //1200chromosome.segment(string)
The chromosome number to draw, e.g. "1" or "X"
Default: 1
chromosome.target(string) id of a div to append the chromosome svg. Specify as string, e.g "#that-div" or a d3 selection Default: the root html document
chromosome.resolution(number) g-band resolutions Default: 550
chromosome.height(number) Height of the chromosome to draw. This is not the total svg height rendered. Default: 17
chromosome.width(number) Total width on the page to render Default: parent's width
chromosome.useRelative(bool)
Render each chromosome relative to their real sizes. Setting this to false will draw the chromosome to the full width.
Default: true
chromosome.showAxis(bool) Display the basepair axis below the chromosome Default: false
chromosome.selectionMode(string) Allow single or multiple selectors on each chromosome. 'multi' or 'single'. To add additional selectors, press and hold the alt/option key while clicking a new band. Default: 'single'
API
cyto_chr.chromosome() Create a new chromosome instance
chromosome.render()
A call to render() will update the svg with the current configurations. If something is changed later, call to render() again to re-draw the chromosome.
chromosome.getSelections() Get an list of all the selections on the chromosome
chromosome.newSelector(start, stop) Create a new selector on the start and stop basepair of the chromosome
chromosome.moveSelectorTo(start, stop) Move the first selector to the start and stop basepair of the chromosome
chromosome.getSVGTarget() Get the current DOM SVG. This is a d3 selection.
chromosome.on(event, callback)
Capture events from user interactions.
Events emmited are: bandclick, selectorchange, selectorend, and selectordelete.
chromosome.on('bandclick', function(e) {
//e contains contextual data
})If the data directory is changed relative to the source, updated it with:
cyto_chr.modelLoader.setDataDir('../data/');Date source:
Data is loaded from static files in the data/ directory. G-banding resolutions included are 400, 550, and 850.
ftp://ftp.ncbi.nlm.nih.gov/pub/gdp/
License
Apache License 2.0