feature-viewer v1.6.0
neXtProt - The knowledge resource on human proteins
This is a code repository for the SIB - Swiss Institute of Bioinformatics CALIPHO group neXtProt project
See: https://www.nextprot.org/
neXtProt feature viewer
The feature viewer is a super easy javascript library to use in order to draw the different features covering a sequence for a better visualization.
Full documentation and live demo : http://calipho-sib.github.io/feature-viewer/examples/
This version is made in Javascript using the D3 library. For the TypeScript version, see : https://github.com/Lisanna/feature-viewer-typescript
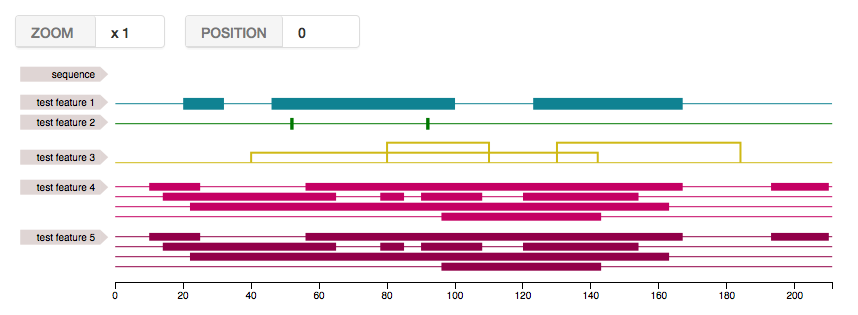
Getting Started
1. You can get the library in your project using NPM/Yarn
//NPM//
npm install feature-viewer
//Yarn//
yarn add feature-viewerOr Include the feature-viewer from jsDelivr CDN in the header of your html
<script src="https://cdn.jsdelivr.net/gh/calipho-sib/feature-viewer@v1.1.0/dist/feature-viewer.bundle.js"></script>NOTE : If you already got the dependencies (D3, Bootstrap & Jquery) in your project, use the simple minified version instead of the bundle :
<script src="https://cdn.jsdelivr.net/gh/calipho-sib/feature-viewer@v1.1.0/dist/feature-viewer.min.js"></script>2. Specify a div in your html
<div id="fv1"></div>3. Create an instance of FeatureViewer in JavaScript with the sequence (or a length), the div in which it will be display and the rendering options of your choice.
var ft = new FeatureViewer.createFeature('MALWMRLLPLLALLALWGPGPGAGSLQPLALEGSLQKRGIVEQCCTSICSLYQLE',
'#fv1',
{
showAxis: true,
showSequence: true,
brushActive: true, //zoom
toolbar:true, //current zoom & mouse position
bubbleHelp:true,
zoomMax:50 //define the maximum range of the zoom
});
//Instead of a sequence, you can also initialize the feature viewer with a length (integer) :
var ft = new FeatureViewer.createFeature(213,'#fv1');To import Feature Viewer into an ES2015 application, you can import specific symbols from specific Feature Viewer modules:
import { createFeature } from "feature-viewer";In Node:
const { createFeature } = require("feature-viewer");4. Finally, add the features
ft.addFeature({
data: [{x:20,y:32},{x:46,y:100},{x:123,y:167}],
name: "test feature 1",
className: "test1", //can be used for styling
color: "#0F8292",
type: "rect" // ['rect', 'path', 'line']
});5. Et voila!
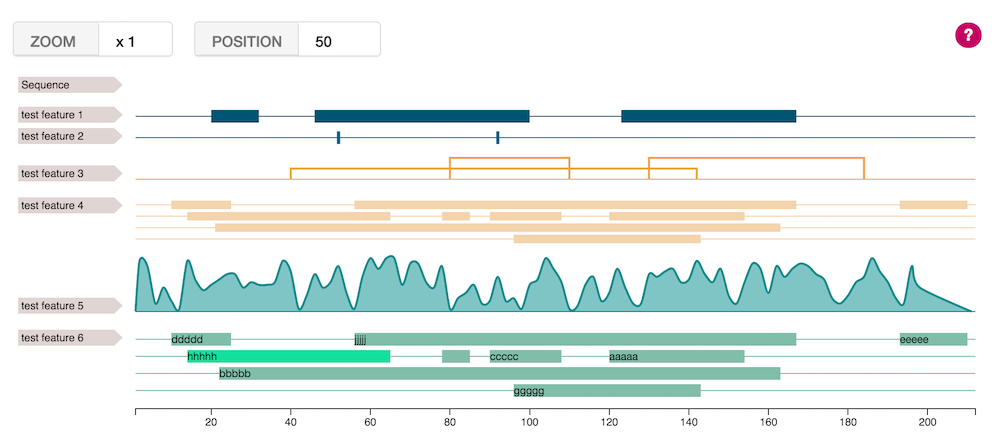
Functionalities
- Zoom into the Feature-viewer by selecting a part of the sequence with your mouse. Zoom out with a right-click.
You can also zoom programmatically with the methodszoom(start,end)andresetZoom()
A tooltip appears when the mouse is over a feature, giving its exact positions, and optionally, a description.
beside the positions for each element, you can also give a description & an ID, allowing you to link click event on the feature to the rest of your project.
Options
- Show axis
- Show sequence
- Brush active (zoom)
- Toolbar (current zoom & position)
- Bubble help
- Zoom max
- Features height
- Offset
ClearInstance()
You may sometimes want to reload your feature-viewer with new parameters. To avoid memory leaks, the method clearInstance() will clear each element & listener for you before you delete the feature-viewer instance.
Documentation
Check out this page for a better understanding of how to use the feature viewer and its possibilities :
Use it with NeXtProt API
It is possible to fill the feature viewer with protein features from NeXtProt, the human protein database.
- First, find your protein of interest in NeXtProt and get the neXtProt accession (NX_...). (You can find your protein by entering an accession number of another database, like UniProt or Ensembl)
- Then, check the type of feature in the NeXtProt API that you would like to add to your viewer. For example, "propeptide" or "mature-protein".
- Include the feature viewer bundle with nextprot to your html : feature-viewer.nextprot.js
- Finally, create your feature-viewer like this :
//initalize nextprot Client
var applicationName = 'demo app'; //please provide a name for your application
var clientInfo='calipho group at sib'; //please provide some information about you
const { nxFeatureViewer, Nextprot } = FeatureViewer;
var nx = new Nextprot.Client(applicationName, clientInfo);
//var entry = "NX_P01308";
var isoform = "NX_P01308-1";
// feature viewer options
var options = {showAxis: true, showSequence: true,
brushActive: true, toolbar:true,
bubbleHelp: true, zoomMax:20 };
// Create nextprot feature viewer
nxFeatureViewer(nx, isoform, "#div2", options).then(function(ff){
// Add first custom feature
ff.addFeature({
data: [{x:20,y:32},{x:46,y:100},{x:123,y:167}],
name: "test feature 1",
className: "test1",
color: "#0F8292",
type: "rect",
filter: "type1"
});
// Add second feature from nextprot
var styles = [
{name: "Propeptide",className: "pro",color: "#B3B3B3",type: "rect",filter:"Processing"},
{name: "Mature protein",className: "mat",color: "#B3B3C2",type: "rect",filter:"Processing"}
];
ff.addNxFeature(["propeptide","mature-protein"], styles);Examples
https://api.nextprot.org/entry/NX_P01308/proteomics
Support
If you have any problem or suggestion please open an issue here.
Development
git clone https://github.com/calipho-sib/feature-viewer.git
npm install (will install the development dependencies)
npm start (will start the development server on localhost:8080)
...make your changes and modifications...
npm run build (will create the bundle js & css in build/)
npm publish (will publish in npm)
License
Copyright (c) 2015, SIB Swiss Institute of Bioinformatics
This program is free software; you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation; either version 2 of the License, or (at your option) any later version.
1 year ago
2 years ago
2 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
3 years ago
4 years ago
4 years ago
5 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
10 years ago
11 years ago
11 years ago
11 years ago
11 years ago
11 years ago
11 years ago