flapjack-bytes v1.0.0
flapjack-bytes
Overview
A JavaScript and HTML5 Canvas based graphical genotype visualization library. The visualization is modelled on our pre-existing Java desktop application Flapjack. Flapjack-Bytes allows you to view graphical gentoype data coloured either by state, or, by similarity to another line in the dataset. You can optionally import a genome map to see a multiple-chromosome view of the data.
The Flapjack-Bytes library support loading data from files on your computer, from URL (has to be CORS compliant, or on the same host), or via BrAPI compliant resources.
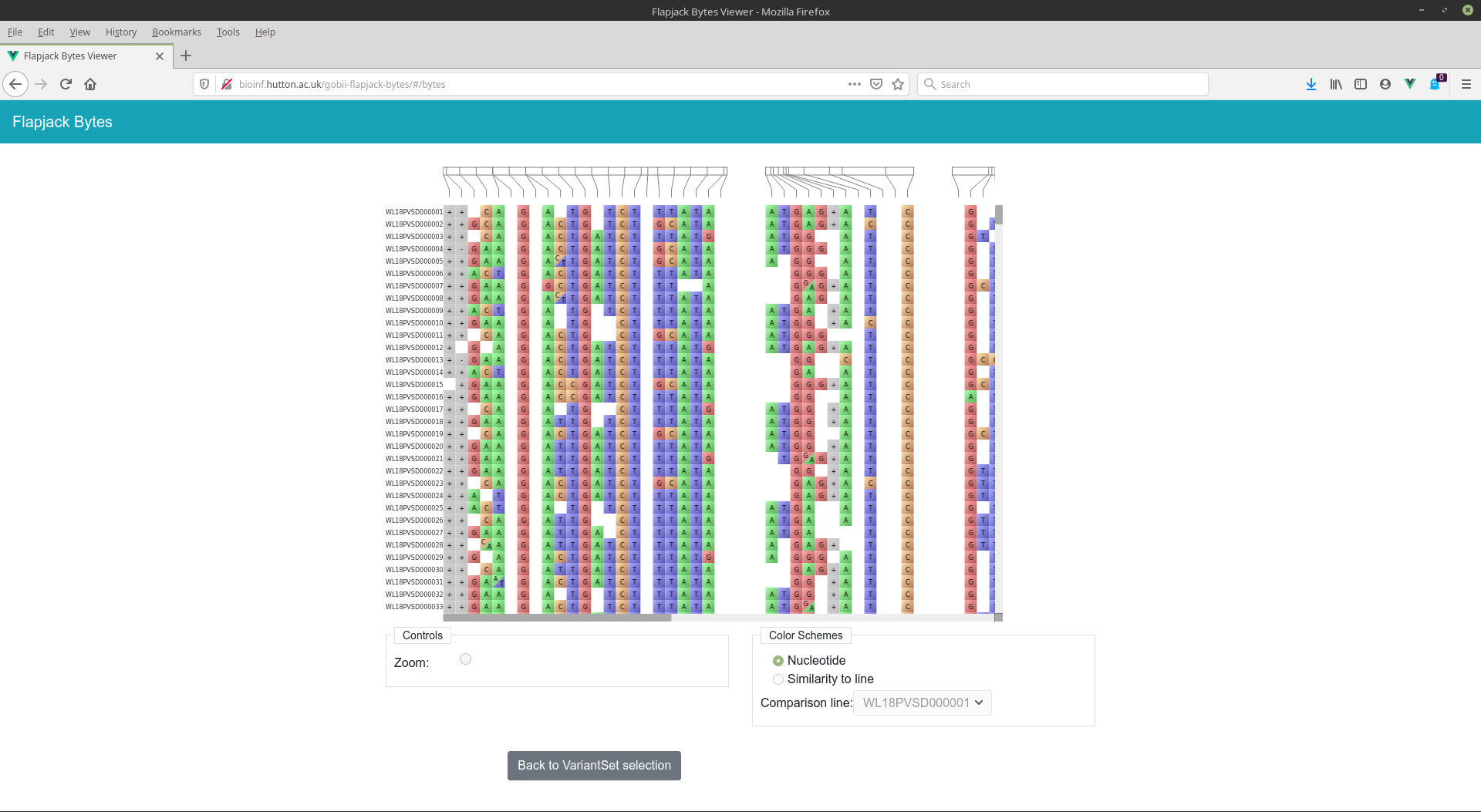
An example of Flapjack-Bytes deployed against the GOBii project's development API can be found at: http://bioinf.hutton.ac.uk/gobii-flapjack-bytes.
Features
- Visualization of genotype data, either in the context of a genomic map, or not.
- Load data from BrAPI enabled resources, files on your local computer, or via URL.
- Multiple colour schemes; the default Nucelotride colour scheme, a similarity colour scheme where each line is coloured on a basis of its match to a user selectable comparison line.
- Highlight of the line and marker under the mouse.
- Zoomable and scrollable view of the data.
Examples
Below are examples of how to integrate the three supported methods of loading data into the library in web pages / web applications.
BrAPI
The Flapjack-Bytes library is designed to work with version 2.0 of the BrAPI specification and doesn't want to be prescriptive about the steps required to search for and select a dataset for viewing. As such the only expectaion is that the calling code will pass Flapjack-Bytes a variantSetDbId and optionally a mapDbId. If a mapDbId is not passed to Flapjack-Bytes, a "fake" map will be constructed and all markers/variants will be displayed on the same chromosome. For BrAPi servers which require authentication, an authentication token can also be passed to the library.
Required BrAPI endpoint
- variantsets/{variantSetDbId}/calls
Optional BrAPI endpoint
- markerpositions?mapDbId={mapDbId}
First define a target div in the web page.
<div id="bytes-div" ref="bytes"></div>The following code is run in the mounted lifecycle hook of the sample Vue app deployed at: http://bioinf.hutton.ac.uk/gobii-flapjack-bytes.
var renderer = GenotypeRenderer();
renderer.renderGenotypesBrapi(
"#bytes-div",
800,
600,
this.baseUrl,
this.callSetId,
this.mapId,
this.authToken
);The baseUrl, callSetId, mapId and authToken are collected on previous pages of the web application and passed to the flapjack-bytes library. mapId can be null if you don't have map data to provide. authToken can be null if the BrAPI resource doesn't require authentication.
The parameters are as follows:
- the target div which we inject the library's canvas into
- the width of the canvas the library will create
- the height of the canvas the library will create
- the base url of the BrAPI server we're communicating with
- the ID of the callSet (genotype dataset) to load from the BrAPI server
- the ID of the map to load from the BrAPI server (or null)
- the authentication token required to talk to the BrAPI server (or null)
Local File
A full example of this can be found in the file load-from-file.html.
<div>
<div>
<label for="mapfile">Map file:</label>
<input type="file" id="mapfile" name="mapfile">
</div>
<div>
<label for="genofile">Genotype file:</label>
<input type="file" id="genofile" name="genofile">
</div>
<input type="submit" action="#" id="submit" name="submit" value="Submit">
</div>
<div id="canvas-holder">
<!-- Empty div that we can insert a canvas into from javascript. -->
</div>The HTML includes a couple of file inputs for getting the files to be loaded into the flapjack-bytes library.
<script src="https://ajax.googleapis.com/ajax/libs/jquery/3.4.1/jquery.min.js"></script>
<script src="build/flapjack-bytes.js"></script>
<script type="text/javascript">
$(document).ready(function(){
$("#submit").click(function(){
var renderer = GenotypeRenderer();
renderer.renderGenotypesFile("#canvas-holder", 800, 600, "#mapfile", "#genofile");
return false;
});
});
</script>Here jQuery is just used to simplify adding the event handler to the submit button. The meat of the code is the following two lines:
var renderer = GenotypeRenderer();
renderer.renderGenotypesFile("#canvas-holder", 800, 600, "#mapfile", "#genofile");Where the parameters are as follows:
- the id of the canvas to inject the flapjack-bytes canvas into
- the width of the canvas
- the height of the canvas
- the id of the file input for the map file (which may or may not have been populated with a file)
- the id of the file input for the genotype file
The map file and genotype file should be in their respective Flapjack formats
Files via URL
A full example of this can be found in the file load-from-url.html.
For this to work the your library integration either needs to be running on the same server that is hosting the files, or the server hosting the files has to be CORS compliant.
<div>
<div>
<label for="mapfile">Map file:</label>
<input type="text" id="mapfile" name="mapfile" value="http://bioinf.hutton.ac.uk/flapjack/sample-data/tutorials/ped-ver-tutorial.map">
</div>
<div>
<label for="genofile">Genotype file:</label>
<input type="text" id="genofile" name="genofile" value="http://bioinf.hutton.ac.uk/flapjack/sample-data/tutorials/ped-ver-tutorial.dat">
</div>
<input type="submit" action="#" id="submit" name="submit" value="Submit">
</div>
<div id="canvas-holder">
<!-- Empty div that we can insert a canvas into from javascript. -->
</div>The HTML includes a couple of text inputs for getting the urls of files to be loaded into the flapjack-bytes library.
<script src="https://ajax.googleapis.com/ajax/libs/jquery/3.4.1/jquery.min.js"></script>
<script src="build/flapjack-bytes.js"></script>
<script type="text/javascript">
$(document).ready(function(){
$("#submit").click(function(){
var renderer = GenotypeRenderer();
renderer.renderGenotypesUrl("#canvas-holder", 800, 600, $('#mapfile').val(), $('#genofile').val());
return false;
});
});
</script>Here jQuery is just used to simplify adding the event handler to the submit button. The meat of the code is the following two lines:
var renderer = GenotypeRenderer();
renderer.renderGenotypesUrl("#canvas-holder", 800, 600, $('#mapfile').val(),
$('#genofile').val());Where the parameters are as follows:
- the id of the canvas to inject the flapjack-bytes canvas into
- the width of the canvas
- the height of the canvas
- the value of the mapfile text input (should be a URL)
- the value of the genotype text input (should be a URL)
7 years ago