genomefeatures v1.0.4
genomefeatures
Screenshot
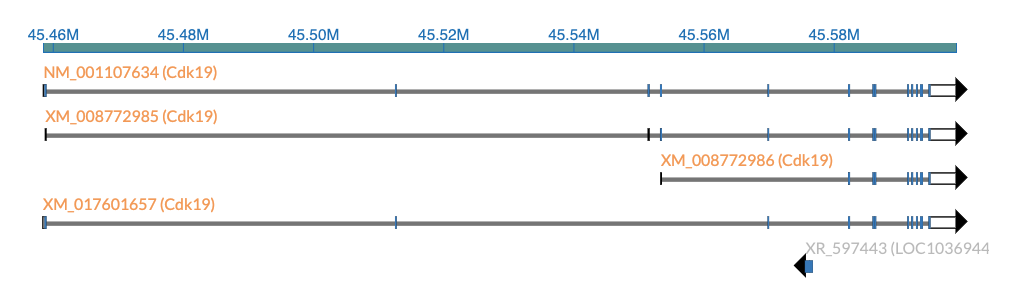
Demo
Demo https://gmod.org/genomefeatures/
Storybook https://gmod.org/genomefeatures/storybook
Instructions
Install from NPM
yarn add genomefeaturesOr see see example/index.html for CDN import style usage, e.g.
Loading data
Legacy access pattern - Apollo REST API
The original implementation of this component required fetching data from an Apollo REST API endpoint. This is still supported, though it is a overkill for most usages
import {
fetchApolloAPIData,
parseLocString,
GenomeFeatureViewer,
} from 'genomefeatures'
const BASE_URL = 'https://www.alliancegenome.org/apollo'
const locString = '2L:130639..135911'
const genome = 'fly'
const region = parseLocString(locString)
const trackData = await fetchApolloAPIFeatures({
region,
genome,
track: 'All Genes',
baseUrl: `${BASE_URL}/track/`,
})
const variantData = await fetchApolloAPIFeatures({
region,
genome,
track: 'Variants',
baseUrl: `${BASE_URL}/vcf/`,
})
const gfc = new GenomeFeatureViewer(
{
region,
genome,
tracks: [
{
type: 'ISOFORM_EMBEDDED_VARIANT',
trackData,
variantData,
},
],
},
`#svgelement`,
900,
500,
)And then in your HTML
<svg id="svgelement"></svg>Static access pattern - Accessing JBrowse NCList files and VCF tabix files
After the refactor, we can now fetch files from static files like JBrowse 1 NCList and VCF tabix files. This means you do not need a complex apollo deployment to use this component: just some static files
import {
fetchNCListData,
fetchTabixVcfData,
parseLocString,
GenomeFeatureViewer,
} from 'genomefeatures'
const locString = '2L:130639..135911'
const genome = 'fly'
const vcfTabixUrl =
'https://s3.amazonaws.com/agrjbrowse/VCF/7.0.0/fly-latest.vcf.gz'
const ncListUrlTemplate =
'https://s3.amazonaws.com/agrjbrowse/docker/7.0.0/FlyBase/fruitfly/tracks/All_Genes/{refseq}/trackData.jsonz'
const region = parseLocString(locString)
const trackData = await fetchNCListData({
region,
urlTemplate: ncListUrlTemplate,
})
const variantData = await fetchTabixVcfData({
url: vcfTabixUrl,
region,
})
const gfc = new GenomeFeatureViewer(
{
region,
genome,
tracks: [
{
type: 'ISOFORM_EMBEDDED_VARIANT',
trackData,
variantData,
},
],
},
`#svgelement`,
900,
500,
)And then in your HTML
<svg id="svgelement"></svg>Developers
git clone git@github.com:GMOD/genomefeatures
yarn dev # vite demo
yarn storybook # storybook examplesNotes
Originally called https://github.com/GMOD/GenomeFeatureComponent
Created by Nathan Dunn (@nathandunn), used by Alliance of Genome Resources
Updated in 2025 by Colin Diesh (@cmdcolin) to add ability to fetch from static files

