ui-tractview v1.0.2
ui-tractview
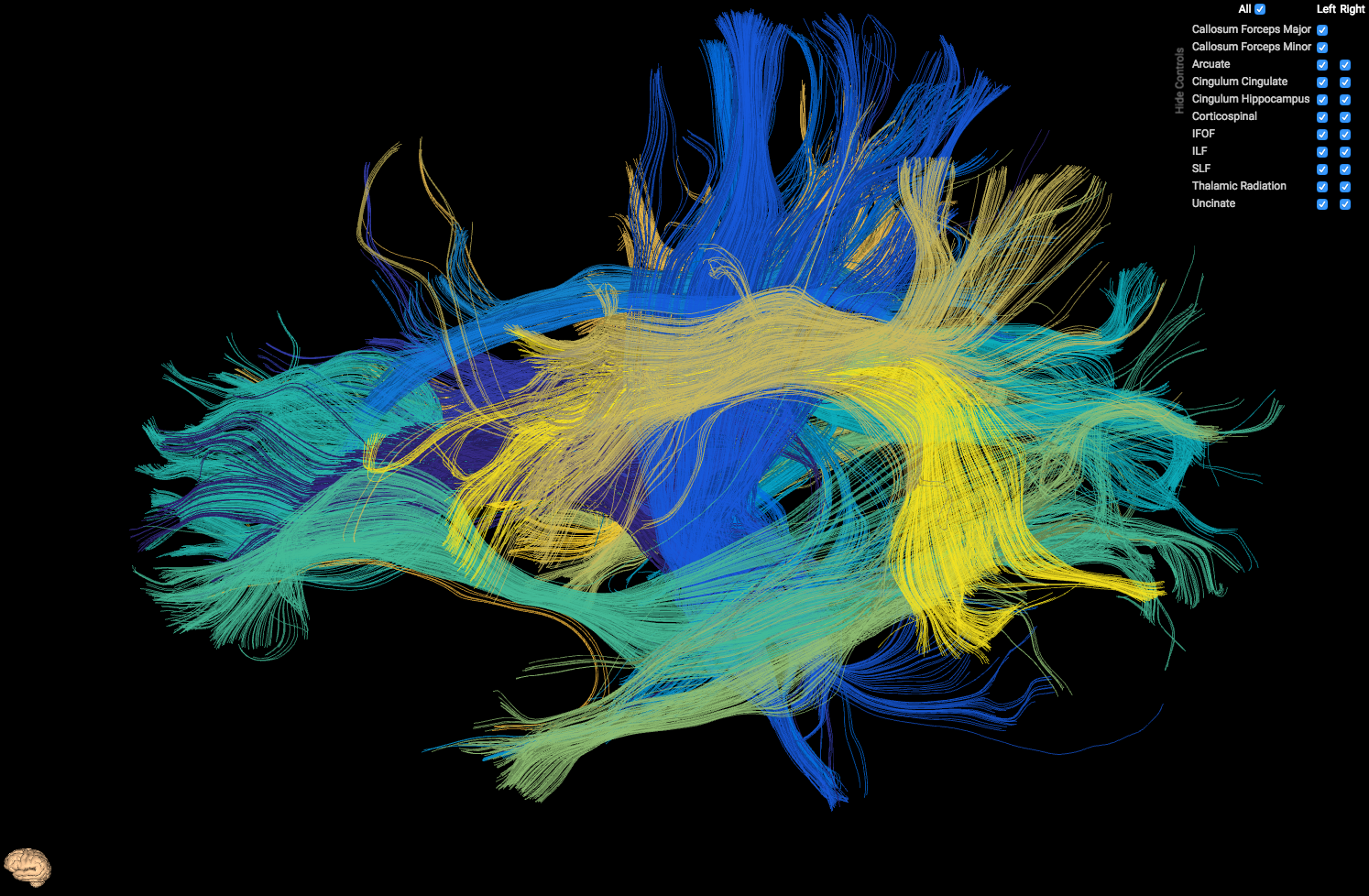
HTML5 Tract Viewer - Used to visualize output from AFQ
Install
Install for general purpose use
npm install ui-tractviewInclude script dependencies in your index.html file:
<!-- Dep Scripts -->
<script type="text/javascript" src="node_modules/jquery/dist/jquery.min.js"></script>
<script type="text/javascript" src="node_modules/three/build/three.min.js"></script>
<script type="text/javascript" src="node_modules/three/examples/js/loaders/VTKLoader.js"></script>
<script type="text/javascript" src="node_modules/bootstrap/dist/js/bootstrap.min.js"></script>
<!-- Main Scripts -->
<script type="text/javascript" src="node_modules/ui-tractview/js/OrbitControls.js"></script>
<script type="text/javascript" src="node_modules/ui-tractview/js/tractview.js"></script>
<!-- Dep Styles -->
<link rel="stylesheet" type="text/css" href="node_modules/bootstrap/dist/css/bootstrap.min.css" />Include your main.js file inside index.html:
<script type="text/javascript" src="js/main.js"></script>
Create an element to append tractview controls to:
<div id='tractviewer' style='position:relative; width:100vw; height: 100vh;'></div>
An explicit width/height for this element is not required, notice that above we just set it to the full window width/height.
Inside main.js, on window load, init the tract viewer:
$(function(){
TractView.init({
selector: '#tractviewer',
num_tracts: 20,
preview_scene_path: 'node_modules/ui-tractview/models/brain.json',
get_json_file: function(tractNumber) {
return 'path_to_json_files/' + encodeURIComponent(tractNumber + ".json");
}
});
});selector represents the query selector for the element that will contain the tract viewer.
num_tracts is the number of tracts that your model contains.
preview_scene_path is the (optional) path to a json scene which displays the orientation of the model. You can either not use previewing by not including this parameter, use ours which is node_modules/ui-tractview/models/brain.json (displayed as the tiny brain in the above image, in the lower left hand corner), or include your own.
get_json_file is a function that takes in a tract number to load, and returns the path to that respective tract. A tract's JSON file should have the following layout:
{
"name": "Left Thalamic Radiation", // If a name begins with Left/Right, it will be respectively prioritized in the tract viewer controls
"color": [0.2081, 0.1663, 0.5292], // [r, g, b] ranging from 0 to 1 each
"coords": [ // list of tracts
[
[-21.69491386, -21.64446831, -21.4675293, ...], // list of x coordinates
[43.13895035, 42.14380264, 41.15979385, ...], // list of y coordinates
[1.224040627, 1.165375113, 1.165637732, ...] // list of z coordinates
],
[
...
],
...
]
}If you are generating an output from AFQ in Matlab, you can use savejson and a script similar to the following in order to create a list of output json files:
[fg_classified,~,classification] = AFQ_SegmentFiberGroups(config.dt6, fg, [], [], false);
tracts = fg2Array(fg_classified);
mkdir('tracts');
cm = parula(length(tracts));
for it = 1:length(tracts)
tract.name = tracts(it).name;
tract.color = cm(it,:);
tract.coords = tracts(it).fibers;
savejson('', tract, fullfile('tracts',sprintf('%i.json',it)));
end